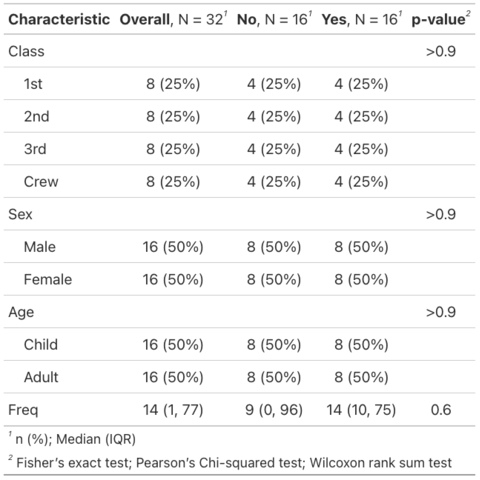
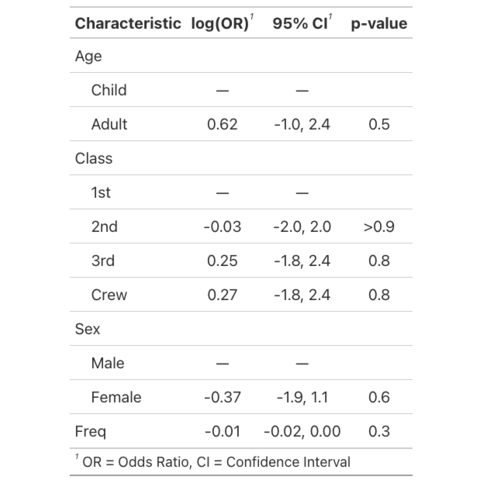
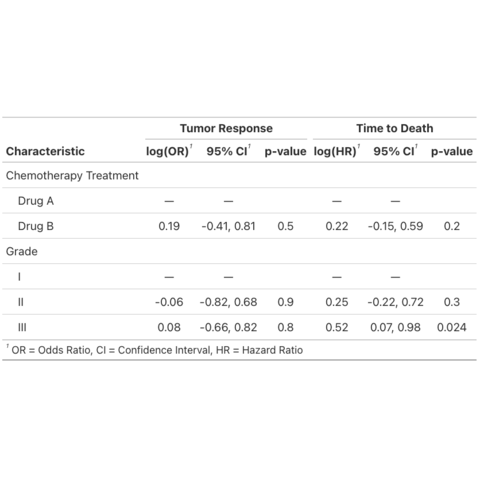
Create summary and presentation ready tables with gtsummary
This post explains how to use the
gtsummary package for creating table summary, especially
with descriptive statistics, regression models, medical values or
demographics data.
This post showcases the key
features of gtsummary and provides a set of
table examples using the package.

{gtsummary}