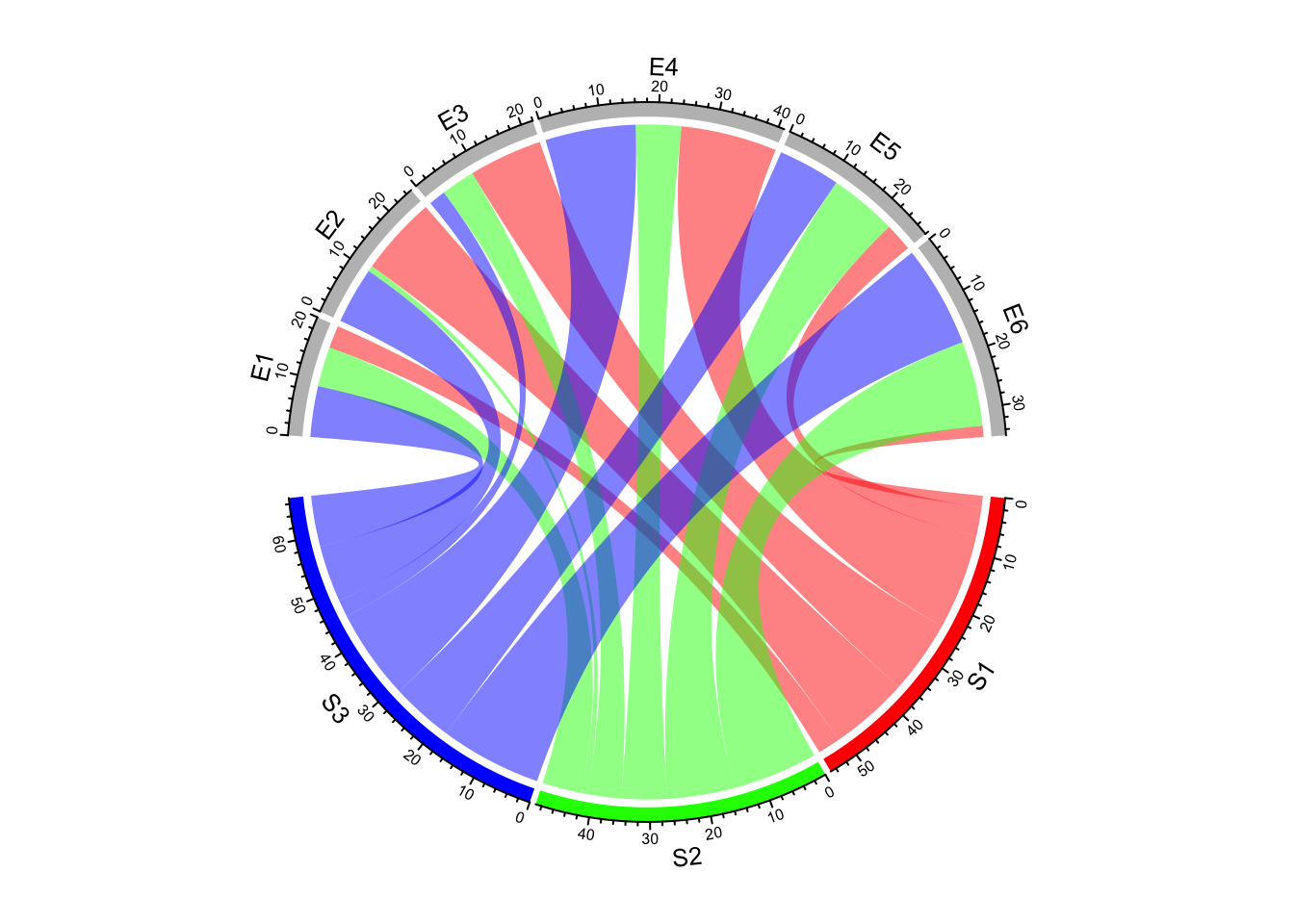
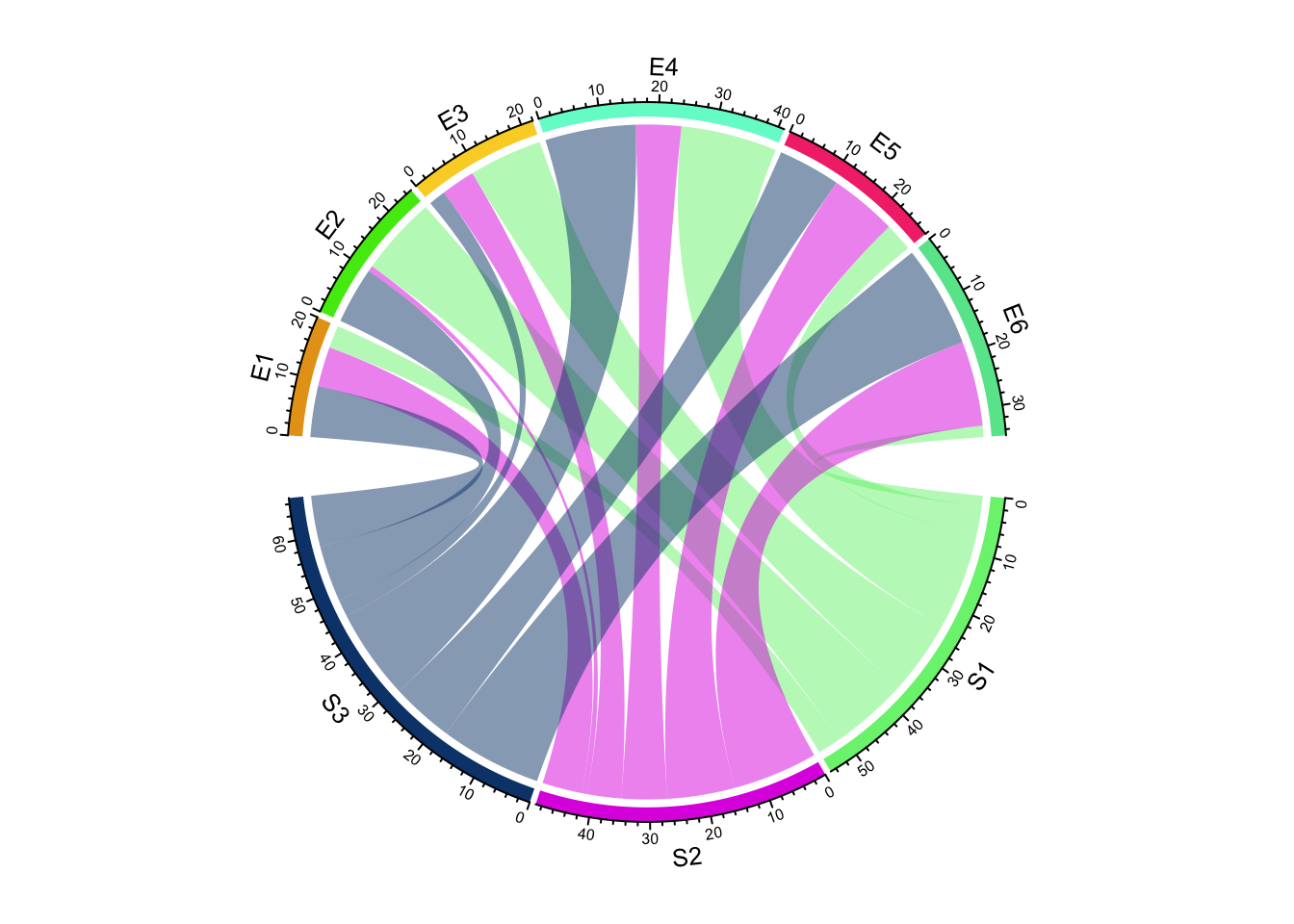
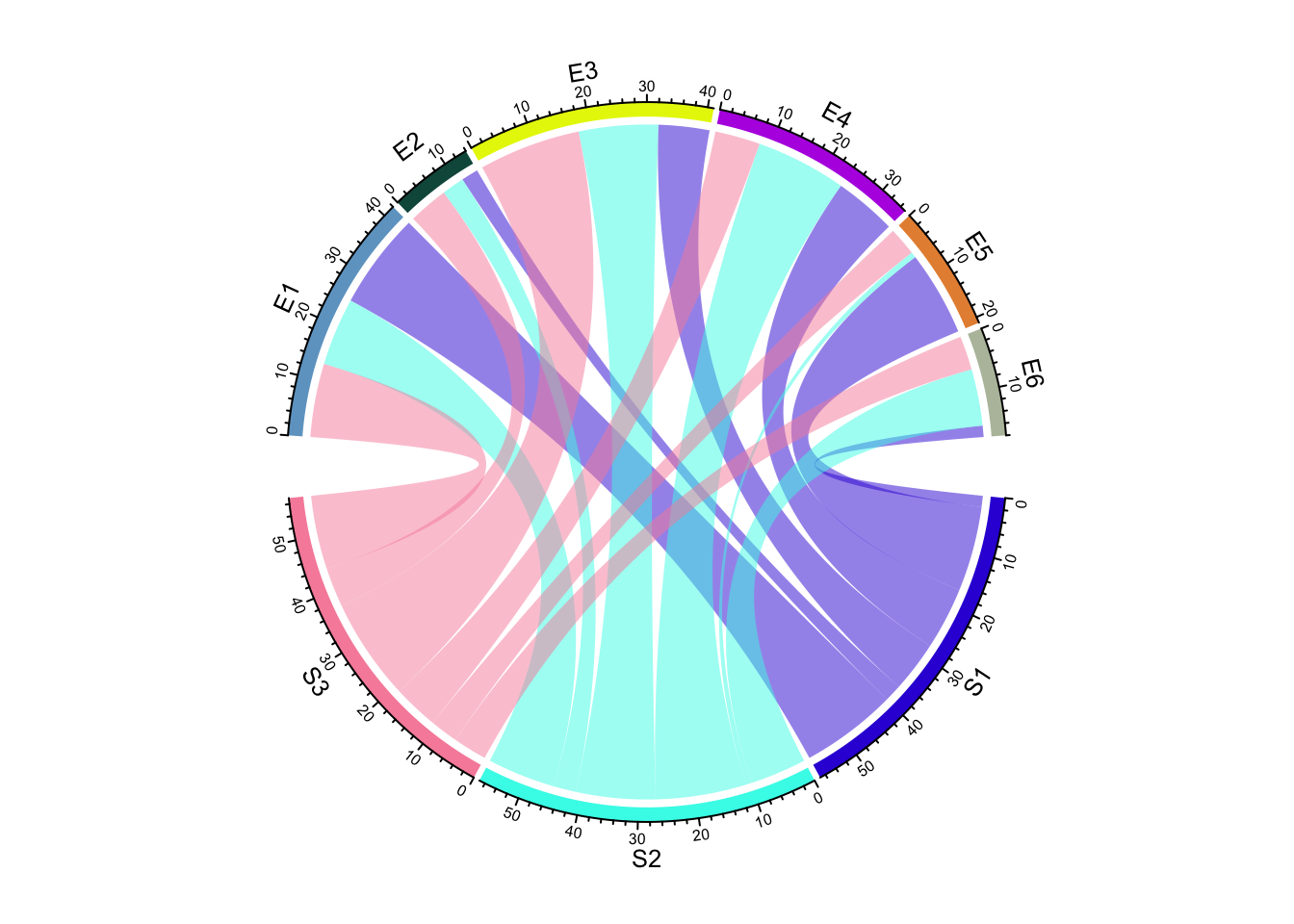
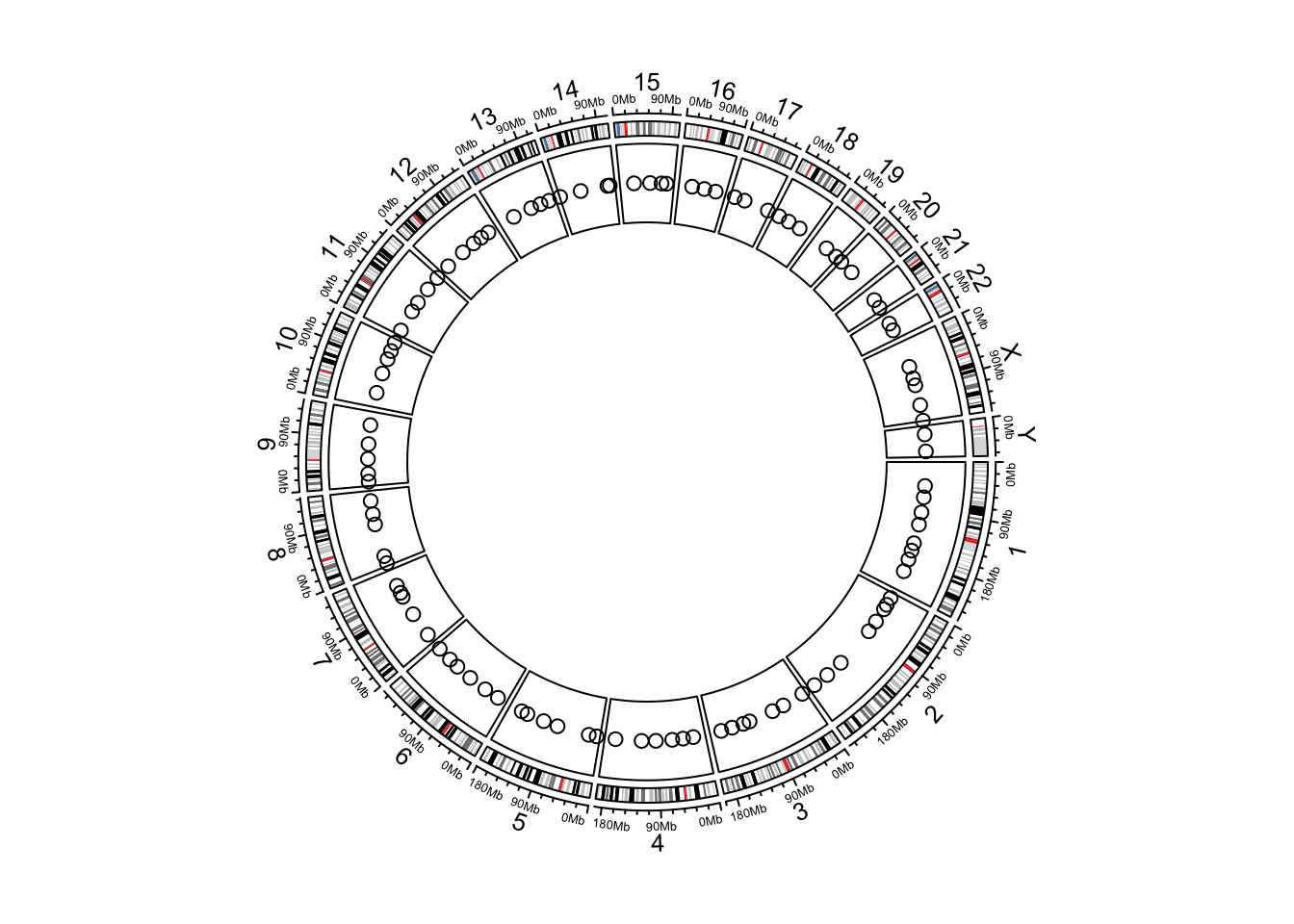
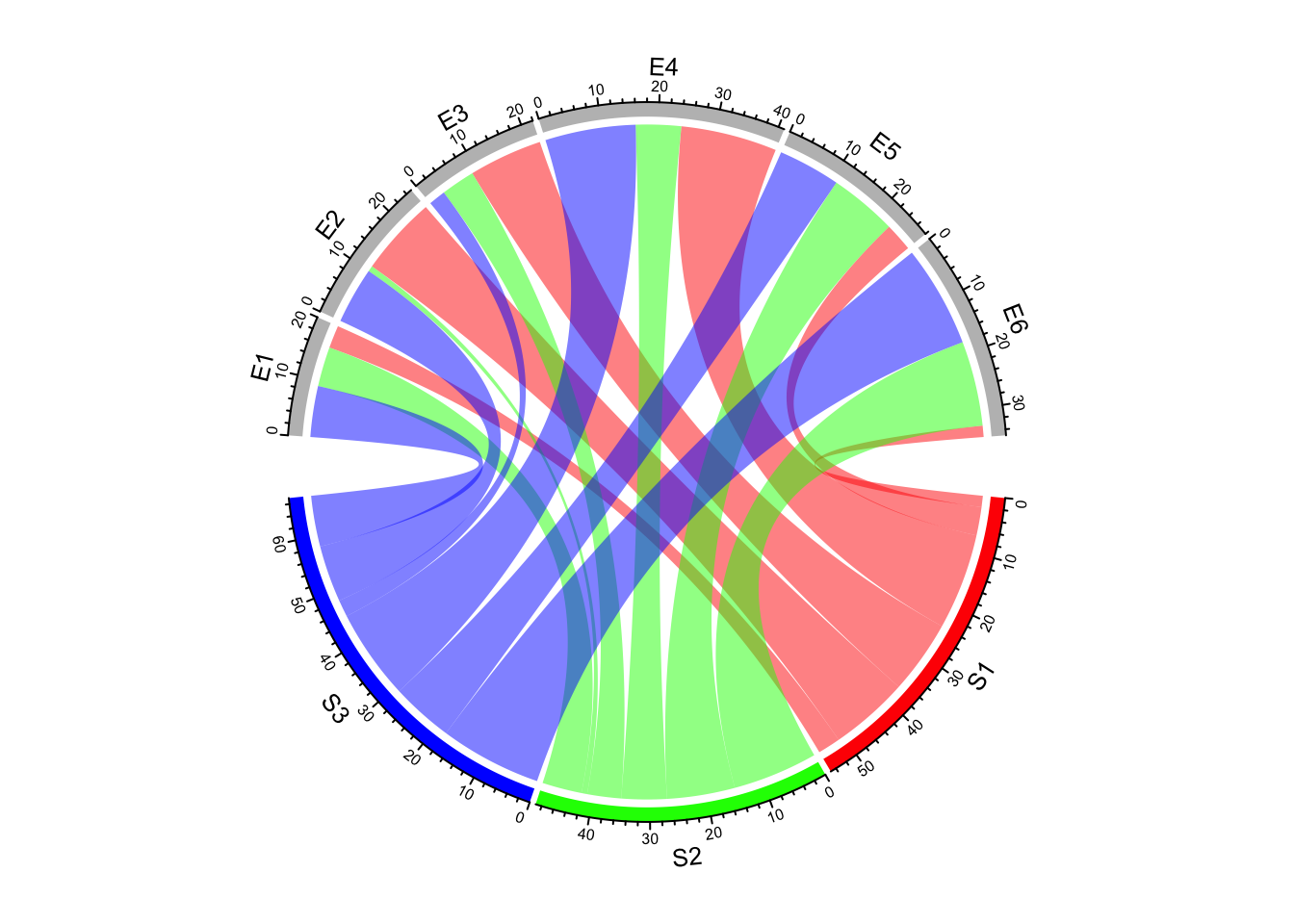
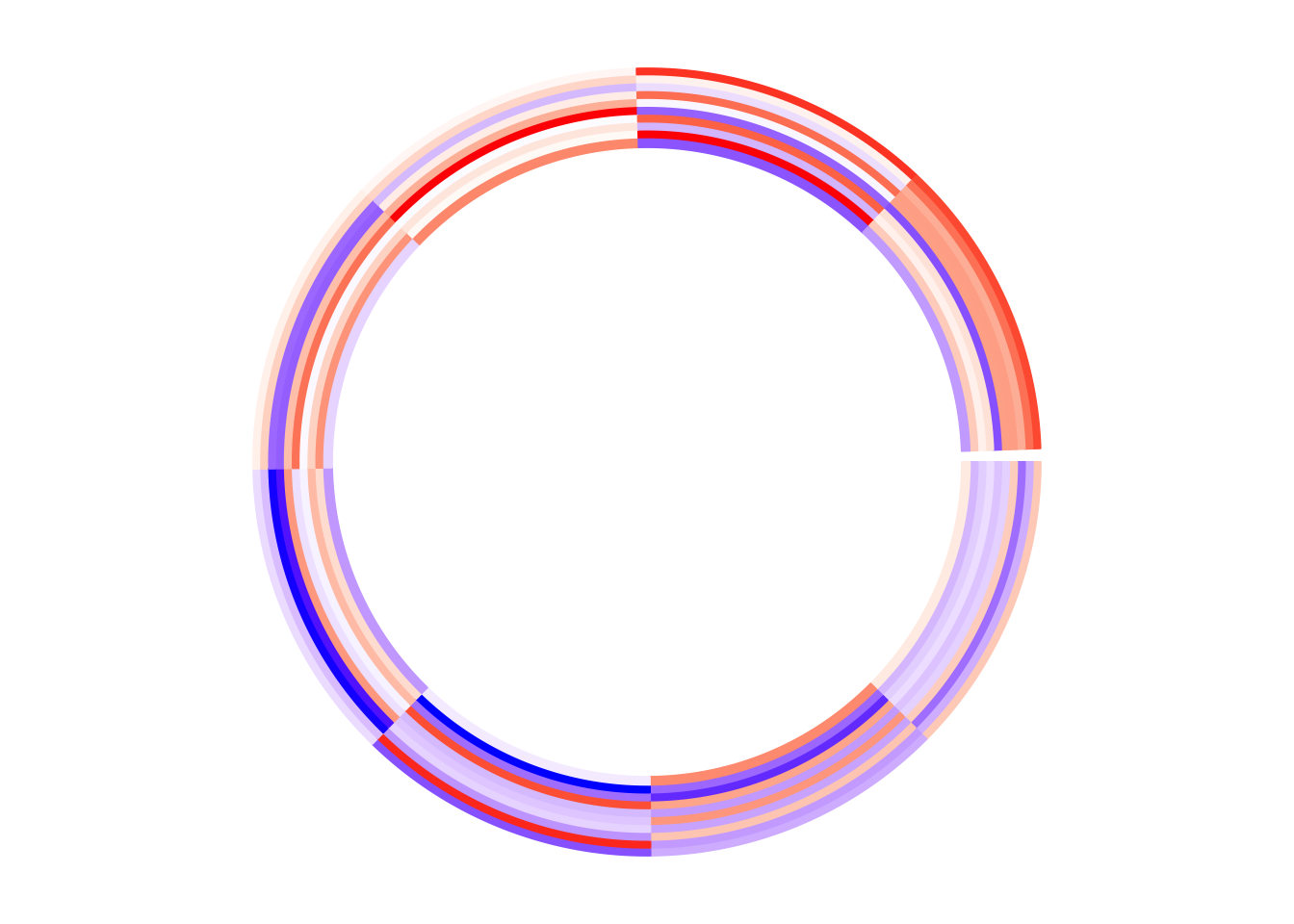
Create Circular Plots with circlize
The circlize package in R is designed for creating
circular plots, including circular heatmaps,
chord diagrams, and more.
This
post showcases the key features of
circlize and provides a set of
graph examples using the package.
{circlize}